The Platforms context contains a set of algorithmic or model-based tools.
After identifying a population of interest, select it in the Analysis tree: Population panel. In the navigation bar, click on the Platforms icon, select which platform you would like to run and the platform window will open in the Discovery panel. (Figure 1)
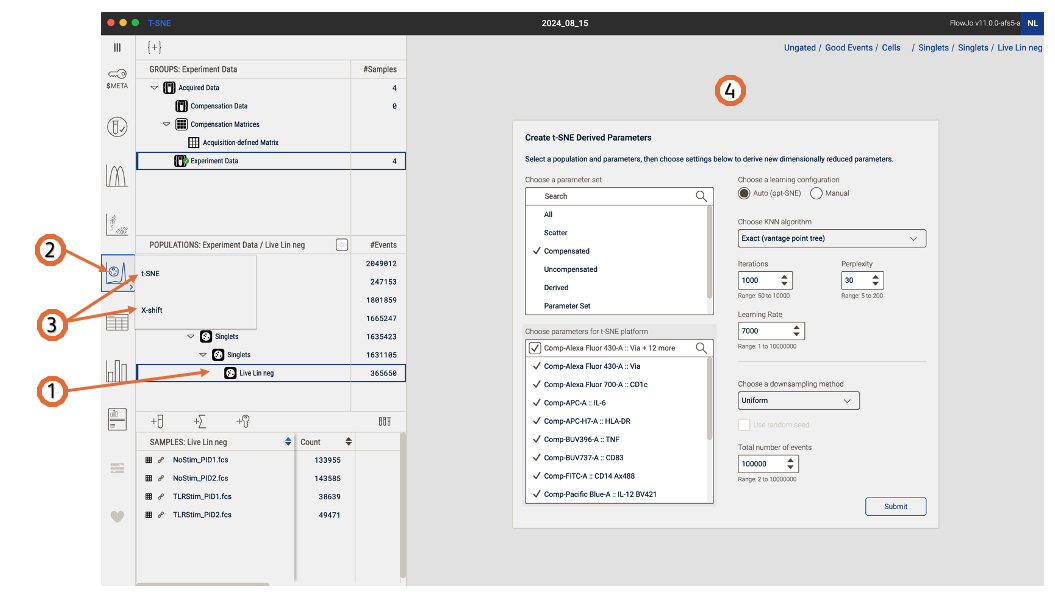
Figure 1. Platforms
t-SNE
T-Distributed Stochastic Neighbor Embedding (t-SNE) is an algorithm for performing dimensionality reduction, allowing visualization of complex multi-dimensional data in fewer dimensions while still maintaining the structure of the data.
t-SNE is an unsupervised nonlinear dimensionality reduction algorithm useful for visualizing high dimensional flow or mass cytometry data sets in a dimension-reduced data space. The t-SNE platform computes two new derived parameters from a user defined selection of cytometric parameters. The t-SNE-generated parameters are optimized in such a way that observations/data points which were close to one another in the raw high dimensional data are close in the reduced data space. Importantly, t-SNE can be used as a piece of many different workflows. It can be used independently to visualize an entire data file in an exploratory manner, as a preprocessing step in anticipation of clustering, or in other related workflows.
How to run t-SNE in v11
After the platform is open, (Figure 2) select the parameters to be used for the t-SNE calculation. You can filter the parameters by choosing a parameter set. If your data is fluorescence-based, make sure to choose only compensated parameters (denoted by the Comp- prefix).
Adjust settings (optional). Defaults have been provided as a starting point and should be acceptable for many data sets. The Opt-SNE algorithm (1), as it utilized an automated Learning Configurations, is highly recommended (and selected by default). Opt-SNE will use the value in the Iterations setting as a maximum, and halt operation when the algorithm stops improving, saving you time. For more information about Opt-SNE check the paper here.
Initiate the calculation by pressing the “Submit” button. The algorithm will run on the input population selected, utilizing selected options. The Platform will create two new parameters, which are the dimension-reduced outputs from the algorithm.
Technical options:
Iterations – Maximum number of iterations the algorithm will run. A value of 300-3000 can be specified.
Perplexity – Perplexity is related to the number of nearest neighbors that is used in learning algorithms. In t-SNE, the perplexity may be viewed as a knob that sets the number of effective nearest neighbors. The most appropriate value depends on the density of your data. Generally, a larger / denser dataset requires a larger perplexity. A value of 2-100 can be specified.
Learning Rate – The learning rate, which controls how much the weights are adjusted at each update. In t-SNE, it is a step size of gradient descent update to get minimum probability difference. A value of 1-10000000 can be specified. Optimally set at 7% the number of cells being mapped into t-SNE space.
KNN algorithm – Sets the k nearest neighbors algorithm. One of the initial steps in t-SNE is to calculate the ‘distance’ or similarity between pairs of cells, using the intensity values of all selected parameters. There are two options, a vantage point tree which is an exact method that calculates all distance between all cells and compares them to a threshold to see if they are neighbors, or the ANNOY algorithm, which is an approximation relying on not necessarily needing all the nearest neighbors to significantly speed up the calculation.
Choose a downsampling method: None, Random or Uniform (For more information about downsampling check this page here) and the Total number of events: Range 2 to 10000000
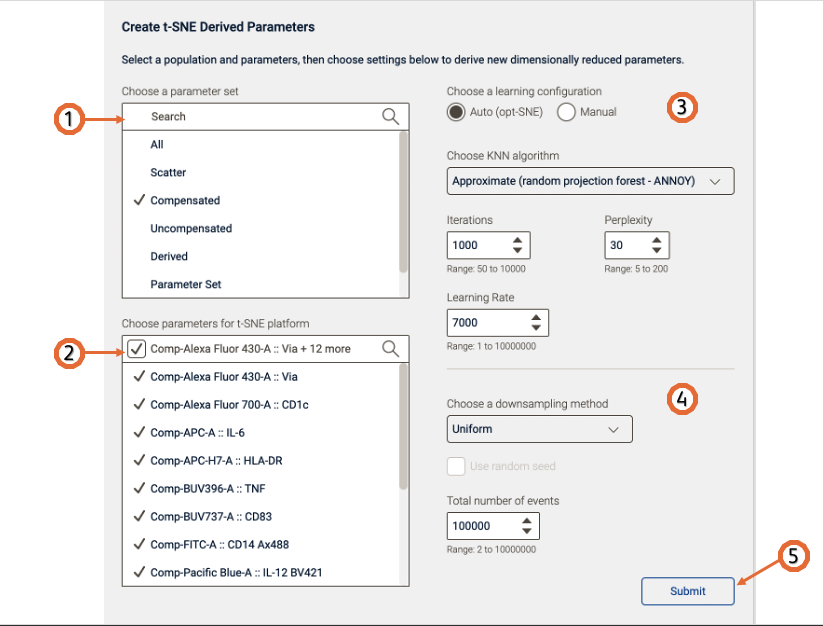
Figure 2. t-SNE platform
X-shift
X-shift (2) is a top-performing clustering algorithm especially suited for discovery of rare cell populations. Another advantage of this unsupervised clustering algorithms vs the other is that it automatically determines the optimal K value.
Within the dialog, select the parameters to use for unsupervised clustering. Once the clustering algorithm finishes calculating it will produce sibling populations as children of the population selected for clustering - These outputs can then be used to further analyze and interrogate the population of interest.
How to run X-shift in v11
After the platform is open (Figure 3), select the parameters to be used for the X-shift calculation. You can filter the parameters by choosing a parameter set. If your data is fluorescence-based, make sure to choose only compensated parameters (denoted by the Comp- prefix).
Adjust settings (optional). Defaults have been provided as a starting point and should be acceptable for many data sets.
Initiate the calculation by pressing the “Submit” button. The algorithm will run on the input population selected, utilizing selected options. The Platform will create the clusters as new populations in the analysis toolbar.
Technical options:
Distance metric - X-shift can work with any distance metric that satisfies the triangle inequality. The default is angular distance where [x – y] represents the angle between vectors x and y.
KNN algorithm – Sets the k nearest neighbors algorithm. One of the initial steps in X-shift is to calculate the ‘distance’ or similarity between pairs of cells, using the intensity values of all selected parameters. There are two options, a vantage point tree which is an exact method that calculates all distance between all cells and compares them to a threshold to see if they are neighbors, or the ANNOY algorithm, which is an approximation relying on not necessarily needing all the nearest neighbors to significantly speed up the calculation. Range 3 to 1000
Choose a downsampling method: None, Random or Uniform (For more information about downsampling check this page here) and the Total number of events: Range 2 to 10000000
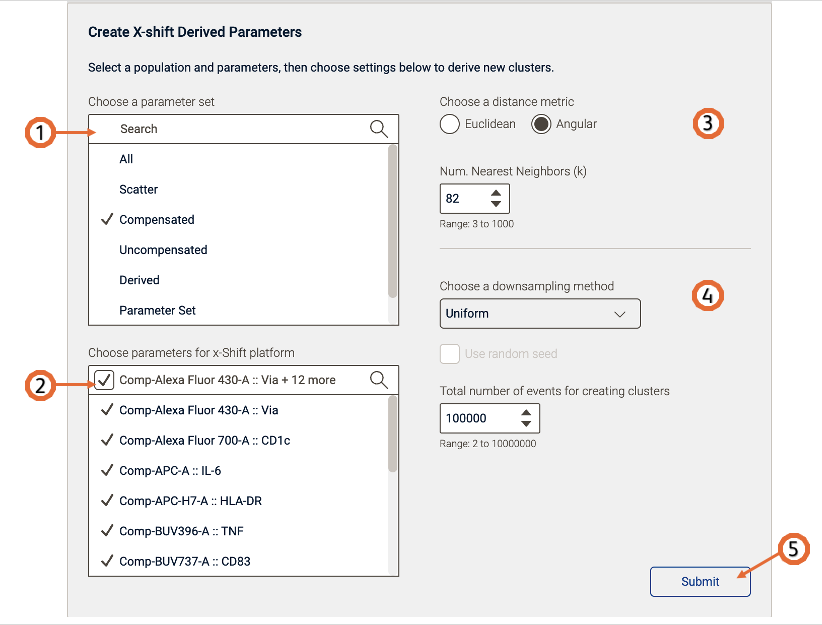
Figure 3. X-shift platform
Belkina, A.C., Ciccolella, C.O., Anno, R. et al. Automated optimized parameters for T-distributed stochastic neighbor embedding improve visualization and analysis of large datasets. Nat Commun 10, 5415 (2019).
Samusik N, Good Z, Spitzer MH, Davis KL, Nolan GP. Automated mapping of phenotype space with single-cell data. Nat Methods.13(6):493-6 (2016).
