is an open source programming environment used by scientists to create software tools for their research needs. Many of these tools have been shared and can be incorporated at no cost into your cytometric data analysis workflow.
is an open source programming environment used by scientists to create software tools for their research needs. Many of these tools have been shared and can be incorporated at no cost into your cytometric data analysis workflow.
There are three ways that you can use R with FlowJo, listed here in order of increasing computer skills.
- Choose from the list of R tools that have been packaged as FlowJo plugins
- Run the R tools from the Gene Pattern server, a web-based tool that uses a largely drag-and-drop or menu based user interface, then import the results into FlowJo for visualization and interaction.
- Run the desired tools directly in R, using a command line interface.
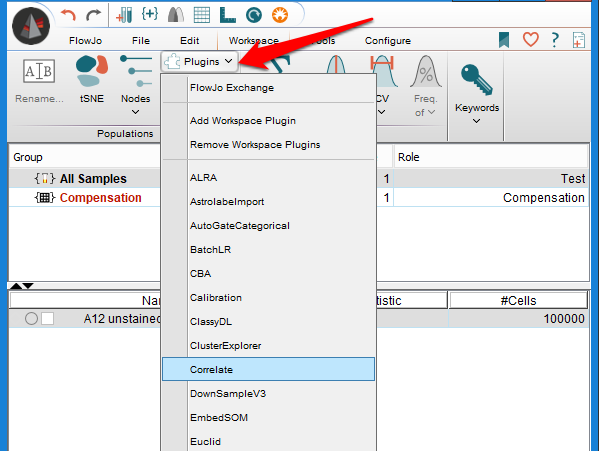
FlowJo Plugins
The FlowJo Exchange hosts plugins, additional tools that can be ‘plugged into’ FlowJo to expand its capabilities. Many of the plugins are were originally developed as R-scripts where they gained fame and popularity. We do our best to stay current and work with the authors of algorithms that are frequently cited or requested by our users to create plugins. All of the plugins are free. R based plugins do not require you to know how to use R, but they do call R in the background to execute the script, and so you must have R installed on your computer. R can be downloaded from http://www.r-project.org/.
Important note as of 8/15/2021: The R versions 4.1.x have significant changes to them that have required many R dependent plugins to be updated. We are in the process of doing this, and you will note that some plugins include a notice that a specific version of the plugin is intended to be run if you have R 4.1.x and an older version of the plugin is needed if you are running R version 4.0.x.
Most R derived plugins require the bioconductor libraries, a suite of R tools developed by scientists working specifically with biological data.
To install the Bioconductor suite, simply open R and type (or copy-paste):
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install()You only need to do this one time. Some R derived plugins will require the user to install the program itself in R (also just one time) before using. This is done through the command ‘install’. As an example, this is the code to install flowSOM, a popular clustering algorithm:
BiocManager::install("flowSOM")
To use plugins in FlowJo you will have to go into the diagnostics tab within the FlowJo plugins and include the R path and the path to whatever folder you have put downloaded plugins in:
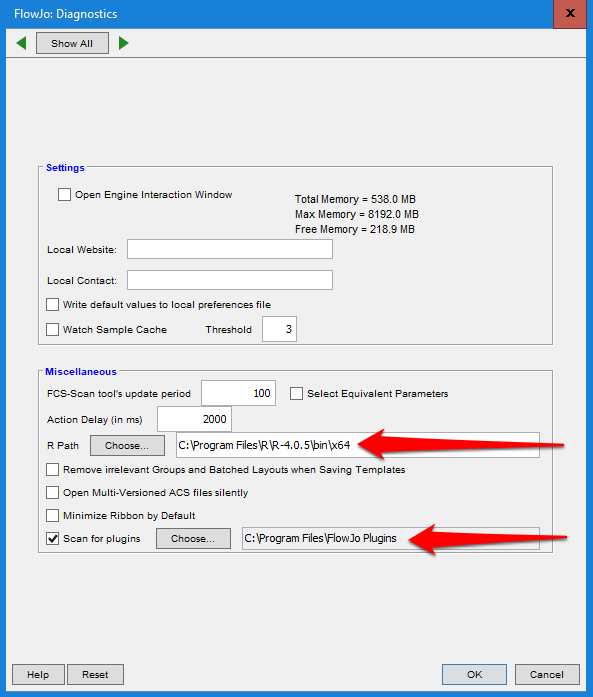
When that has been set up, every plugin you download and drop into the folder you selected will appear in the workspace plugin dropdown menu:
Run the R tools from the Gene Pattern Server
Many of the tools in bioconductor have been made available to non-programmers by the Gene Pattern Server, a group based out of MIT’s Broad Institute. They create a web browser interface that only requires the user to answer a few questions to run the algorithm.
The gene pattern server can be accessed online. You will need to create a user account, but its free. The picture below shows a screen shot of a the gene pattern server with the clustering modules section opened. You can choose from several categories through the ‘Browse Modules’ option, and drag favorites to the selection box to the left.
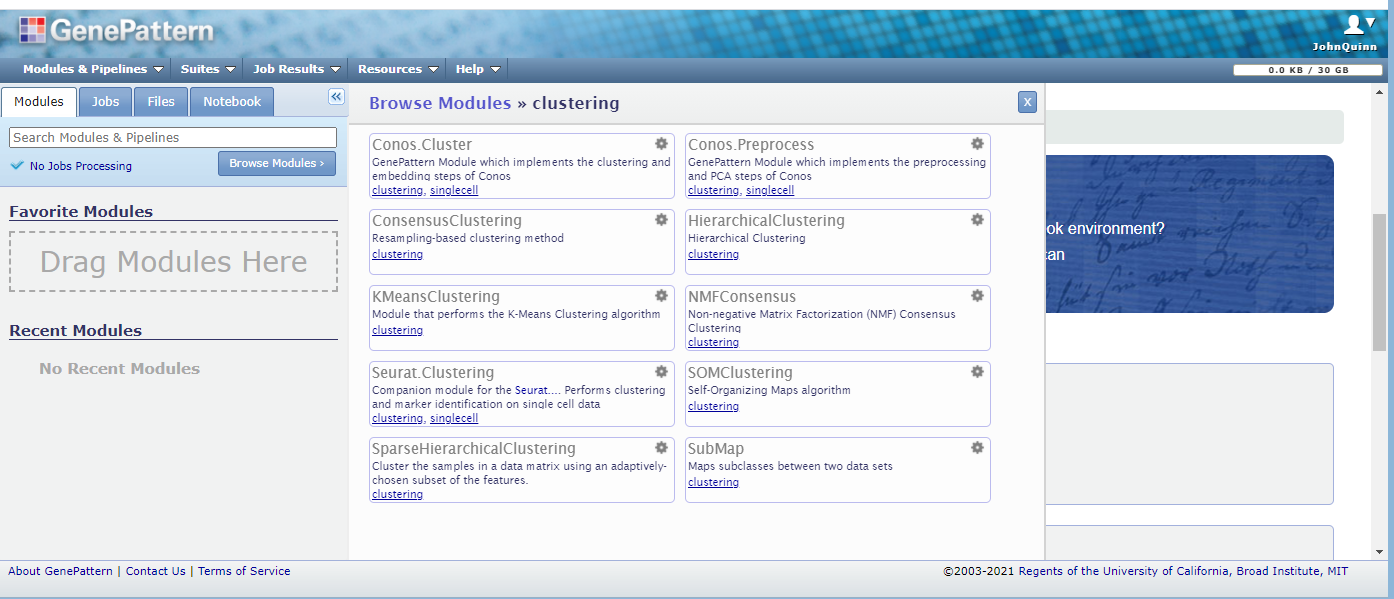
The results are then exported depending on the type of application. For clustering applications the results are usually available as comma separated value (CSV) files, which usually outputs a vector of cluster memberships separated by commas that programs like Excel or FlowJo can interpret. To get these results into FlowJo, drag and drop the resulting CSV file onto the FCS file in FlowJo that you clustered. The event numbers must match, but this should be the case if it is the same file.
Run FlowJo and the desired R tools in a script from the command line
If your are facile with R, you can of course run the R algorithms directly in R and write the outputs as .csv files. As is the case with using the Gene Pattern server, clustering outputs or other derived parameters can be appended to files in FlowJo via drag and drop onto the original file in FlowJo.