Introduction
The FlowJo plugin for ezDAFi bridges automated clustering techniques and human-driven gating strategies. DAFi stands for Directed Automated Filtering and identification of cell populations in polychromatic flow cytometry data. The major feature of ezDAFi is its ability to control unsupervised clustering tools for the identification of previously known populations of interest in high-dimensional flow cytometry datasets. Among other many advantages, ezDAFi-derived gating results follow a much more natural distribution of the data. For more details, please see: https://doi.org/10.1002/cyto.a.23371.
Under the hood, the FlowJo plugin extracts the manual gating strategy from FlowJo to set the boundaries
of regions of interest in the parameter hyperspace. Then, it gates on learned cluster centroids located in the regions of interest and finds all cells whose nearest neighbors are centroids of interest. Finally, the plugin automatically gates each cell according to their updated identities, returning them to the FlowJo workspace.
On a Mac, please install XCode and XQuartz. Open the Terminal and run the following command:
xcode-select –install
The plugin will attempt to auto-install the R dependencies when initiated. To manually install the R dependencies, open R and install the Rcpp and glue packages using:
install.packages(pkgs = c(“Rcpp”, “glue”), repos = ‘http://cran.us.r-project.org’)
On Windows, you will also need to install Rtools IMPORTANT when installing Rtools, you will want to make sure that the box is checked to have the Rtools installer edit the system PATH. If your User account does not have Administrator rights, you may need help from your IT department to install.
There are detailed instructions on how to set up plugins in our online documentation.
The ezDAFi plugin can be set up with the following steps:
Place the ezDAFi JAR file into a “plugins” folder.
1. Ensure that the R program is installed on the computer.
2. Go to the Preferences (heart) icon in FlowJo and select Diagnostics.
3. Select the plugins folder.
4. Set the correct R path in the R Path field.
5. Restart the application.
Using the ezDAFi plugin.
The plugin automatically imports single-cell data and FlowJo-derived gating strategies into R, builds single cell representations with either self-organizing maps (SOM, preferable) or k-means, and updates single cell identities according to the location of their nearest cluster centroid within the gating tree. The plugin can be used to refine gated populations in FlowJo, allowing for the visual check and re-use of ezDAFi gated cells in downstream analysis, such as building stats and plots.
While building the gating tree, the user can select a parent population, call the plugin and refine all of its
child populations using ezDAFi.
NOTE: You must save the workspace before running this plugin – each time the plugin is run.
This ensures the plugin gets the most up-to-date gating hierarchy available in the workspace, and also the most up to date list of samples in the workspace.
To start the plugin, first select a population in the workspace that has at least one child population, then
select the ezDAFi plugin from the plugin drop-down menu. You will be presented with several options in
the ezDAFi dialog window.
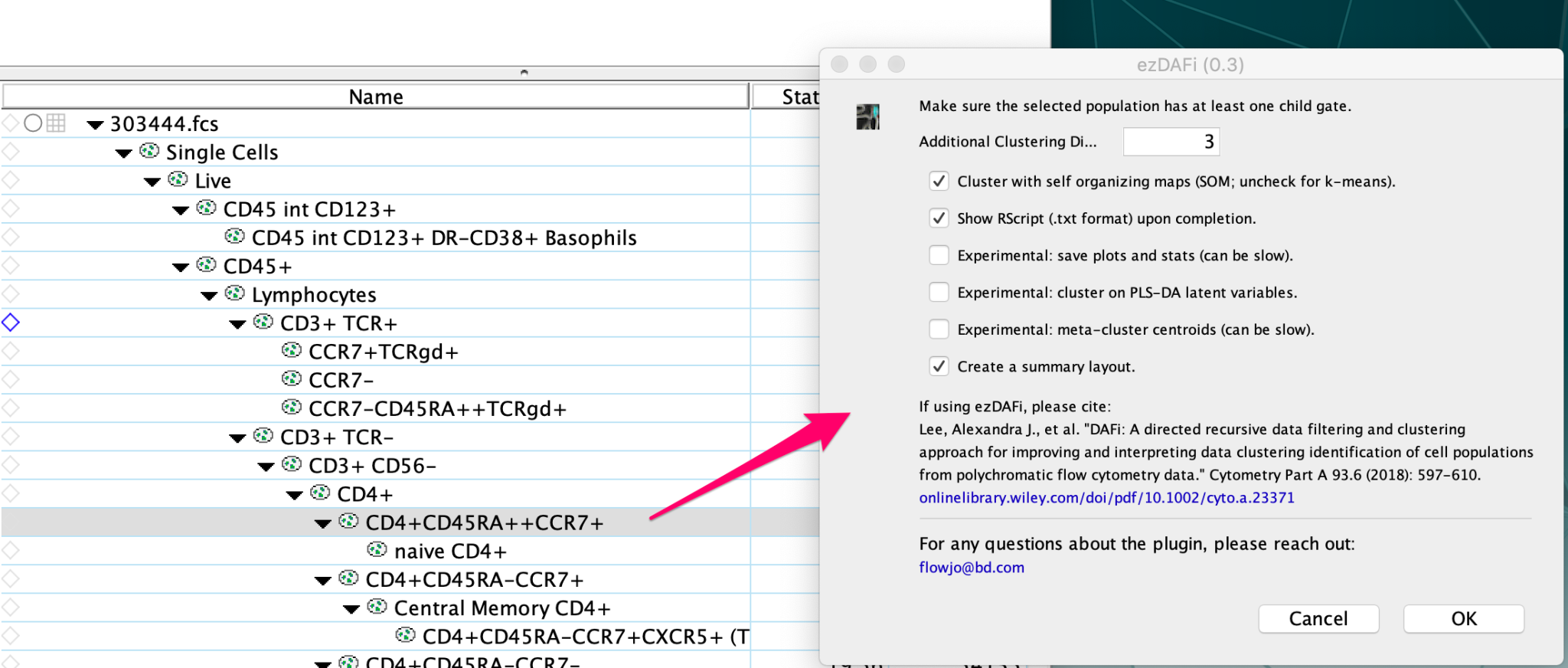
Leaving the first option selected will allow for refinement of your populations with self-organizing
maps (recommended) while un-checking this option will instead use k-means clustering for population
refinement.
• Leaving the second option selected will show the Rscript on screen when the plugin completes.
• Selecting the third option to save plots and stats will allow the plugin export a heatmap comparing
your manually gated populations to the ones refined by ezDAFi across all parameters. You will also
get pseudocolor plots of your manually gated populations compared to the ezDAFi population as well
as .csv files of counts and frequency of parent statistics.
• Selecting the fourth option to cluster on PLS-DA latent variables will allow the plugin to run partial
least squares discriminant analysis on the data before clustering.
• Selecting the fifth option to meta-cluster centroids can help stabilize gating results when there is
technical variation in the data.
• Leaving the sixth option selected to create summary plots will allow FlowJo to create overlays in the
Layout Editor of the ezDAFi populations on top of the parent population.
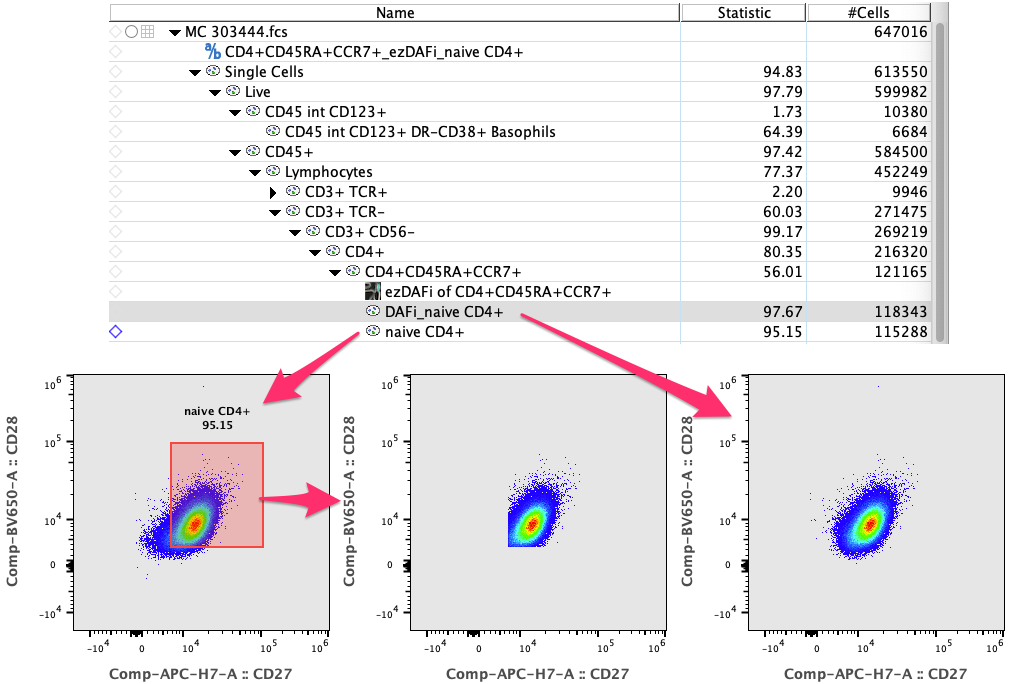
Figure 1: Selecting “CD4+CD45RA+CCR7+” and running the plugin will refine all gates down the hierarchy from “CD4+CD45RA+CCR7+”. In this case, we only show one example of one immediate child gate, the “naive CD4+”. Please note how the ezDAFi-refined population shows a much more natural distribution of the parameters after gating.
The user can also build the whole gating tree first and then apply ezDAFi to a upstream population for the plugin to refine all children and children of children of that population. This feature allows for ezDAFi to be used recursively throughout several levels of the gating tree without additional human interaction. This way, all sub-populations downstream of the selected gate will be refined by ezDAFi.
Leave us your feedback
Please write to flowjo@bd.com with any questions!
References
Lee et al. DAFi: A directed recursive data filtering and clustering approach for improving and interpreting
data clustering identification of cell populations from polychromatic flow cytometry data
https://doi.org/10.1002/cyto.a.23371.
https://github.com/PedroMilanezAlmeida/ezDAFi

