Introduction
X-Shift is a top-performing clustering algorithm especially suited for discovery of rare cell populations. New, updated implementation automatically determines the optimal K value and employs FLANN library to dramatically speed up the KNN search, offering 5X improvement over the original implementation.
Developed and Maintained by Nikolay Samusik
Algorithm Invented by Nikolay Samusik
Installation
After downloading the plugin, place the JAR file into your FlowJo plugins folder and restart FlowJo.
Workflow Overview
After identifying a population of interest select it and run the plugin. The plugin will perform clustering in the population selected.
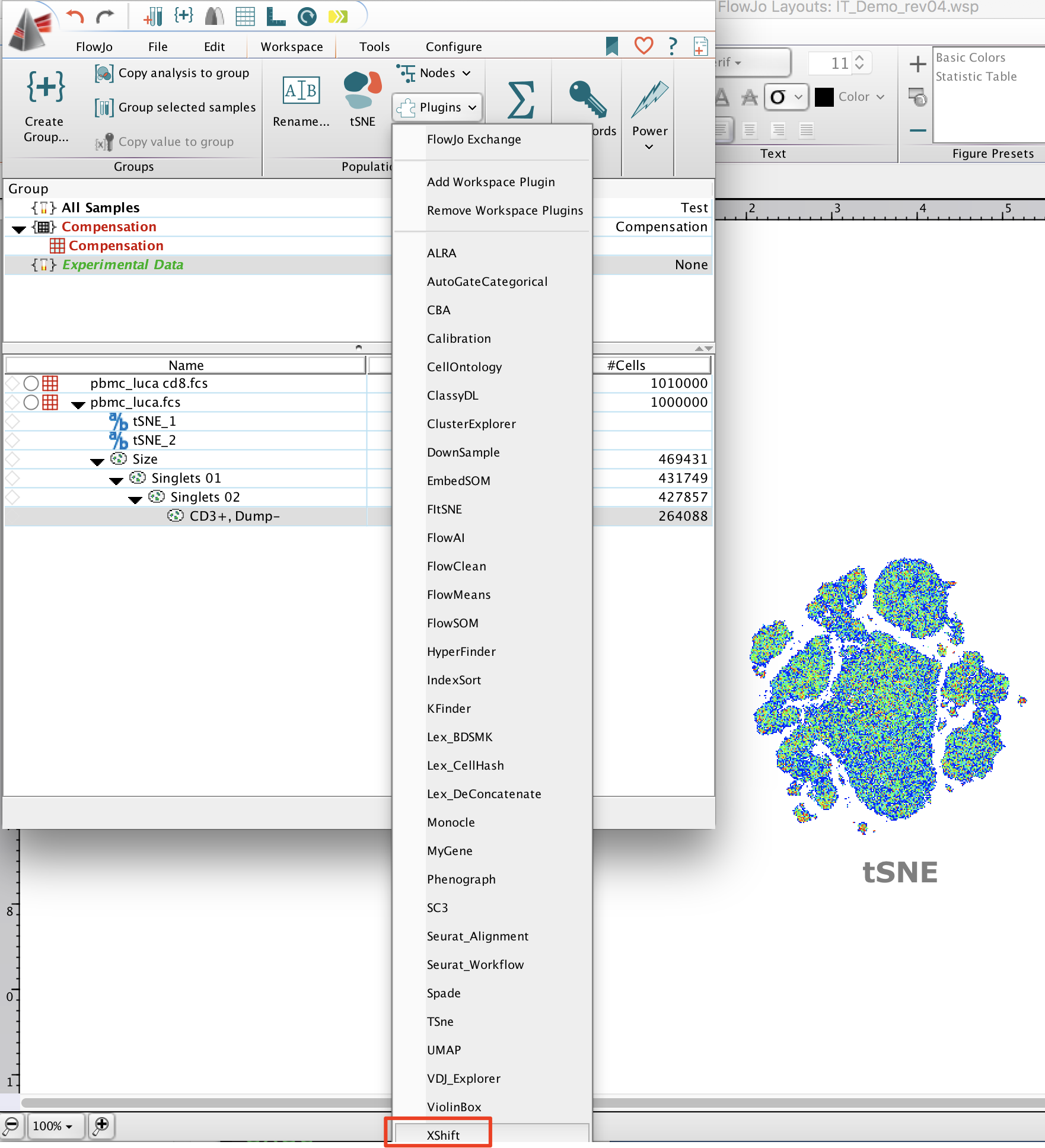
Note: Here we’re performing X-Shift clustering on T Cells from PBMCs stained with cytokines.
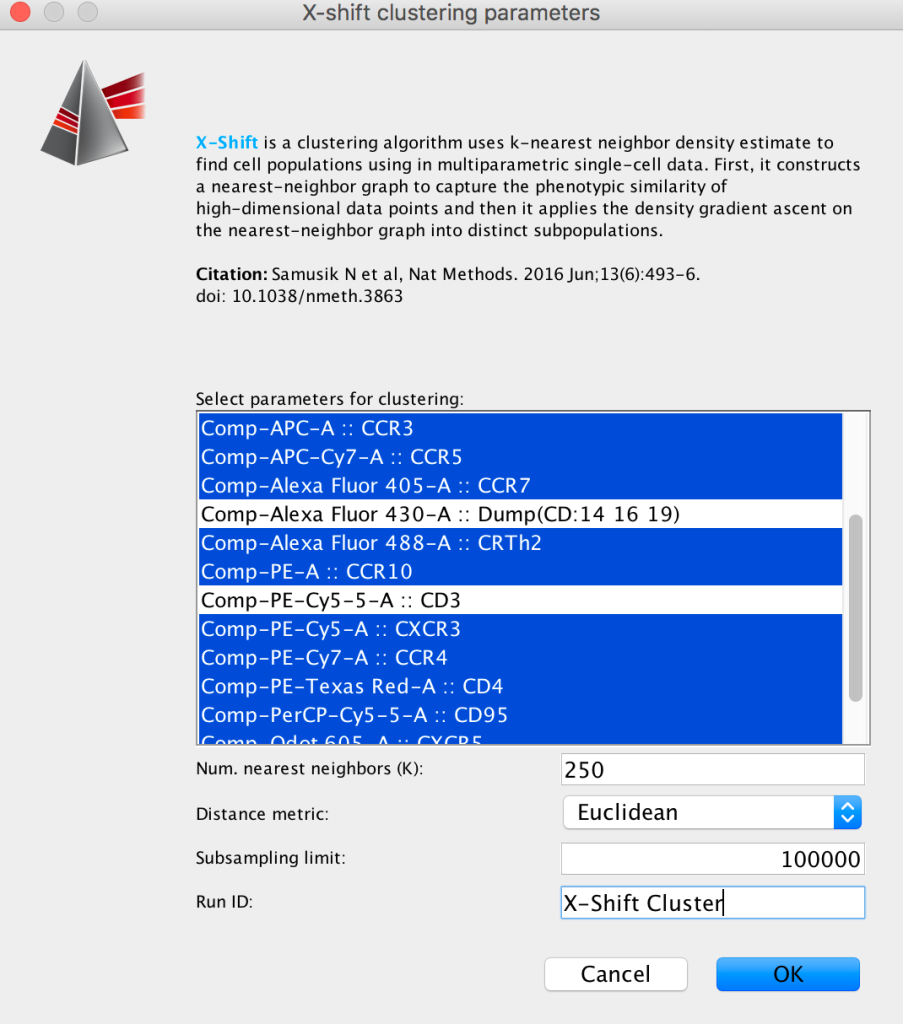
Within the plugin dialog, select the parameters to use for unsupervised clustering.
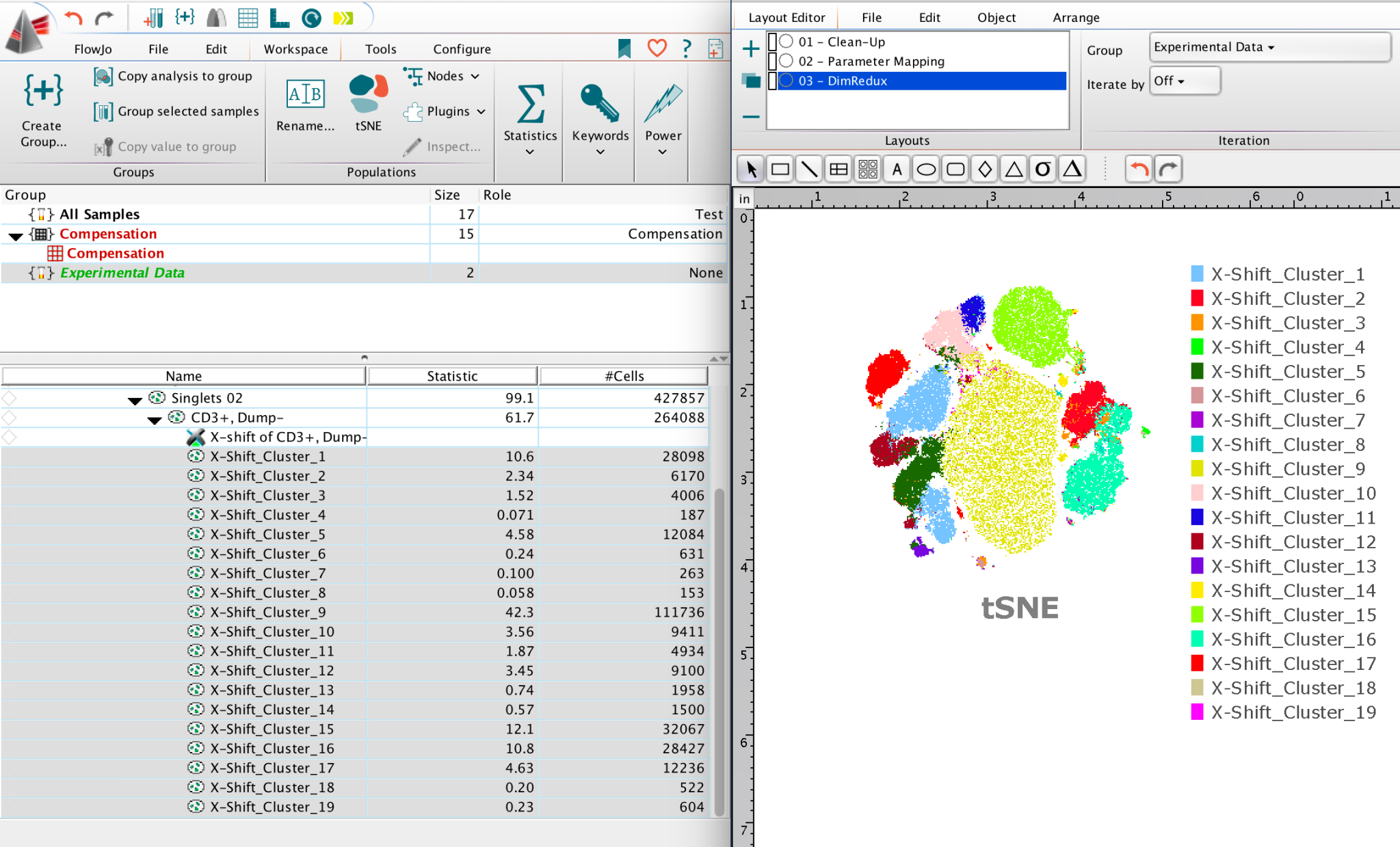
Once the clustering algorithm finishes calculating it will produce sibling populations as children of the population selected for clustering – These outputs can then be used to further analyze and interrogate the population of interest.
Benchmarking
On a system conforming to recommended specifications (16Gb RAM, quad-core processor) – Clustering time for 0.25M cells, KNN = 250, and 12 parameters took 32 minutes.
Support
As you have further questions regarding the use of this tool or other platforms in FlowJo, please reach out: flowjo@bd.com
Our thanks go out to Tess Brodie and her team at the Institute for Research in Biomedicine, Cellular Immunology, Bellinzona for generously providing their data to the public for analysis and review – PBMC data illustrated in this documentation is freely available form Flow Repository:
